Our research is based on the ability of bacteria to quickly acquire new genetic material through horizontal gene transfer, allowing evolutionary timescales that can be measured in minutes rather than eons.
Our basic studies include:
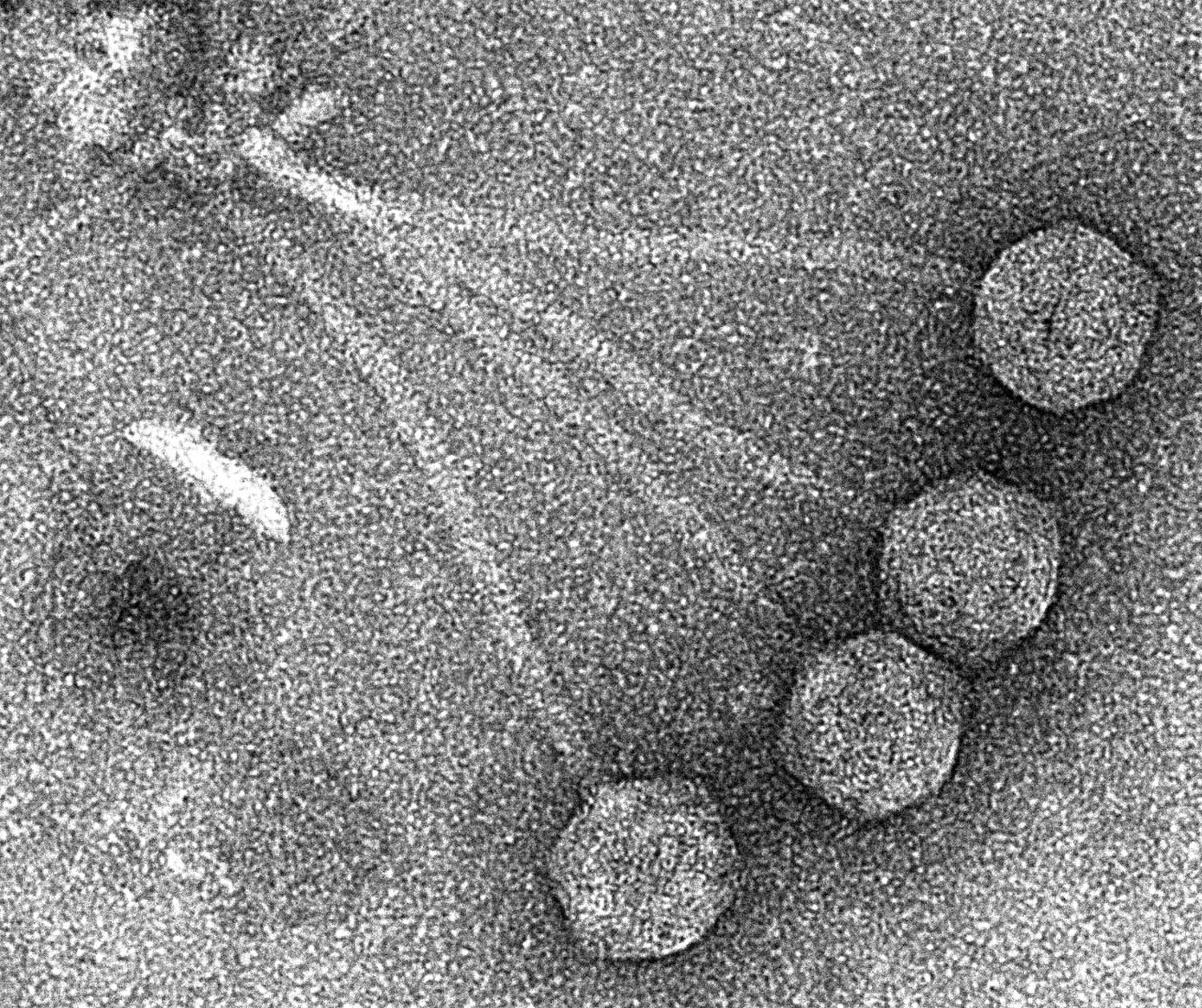
CRISPR and phages
in Streptococcus pyogenes
Cas9 from S. pyogenes has become the prototypical CRISPR system for use in genome editing. However its function as an adaptive immune system to protect against phage infection in S. pyogenes has remained understudied. Characterizing the dynamic between CRISPR and S. pyogenes phages will not only shed light on Cas9 function, but also on the pathogenicity of this bacterium, as integrated phages are major contributors to S. pyogenes virulence. Defining this will address important outstanding questions related to the mechanism and regulation of HGT in S. pyogenes: How does CRISPR immunity interact with the phages of S. pyogenes? How do these phages contribute to pathogenesis? Can these interactions be used to manipulate Cas9 in a genome editing context?
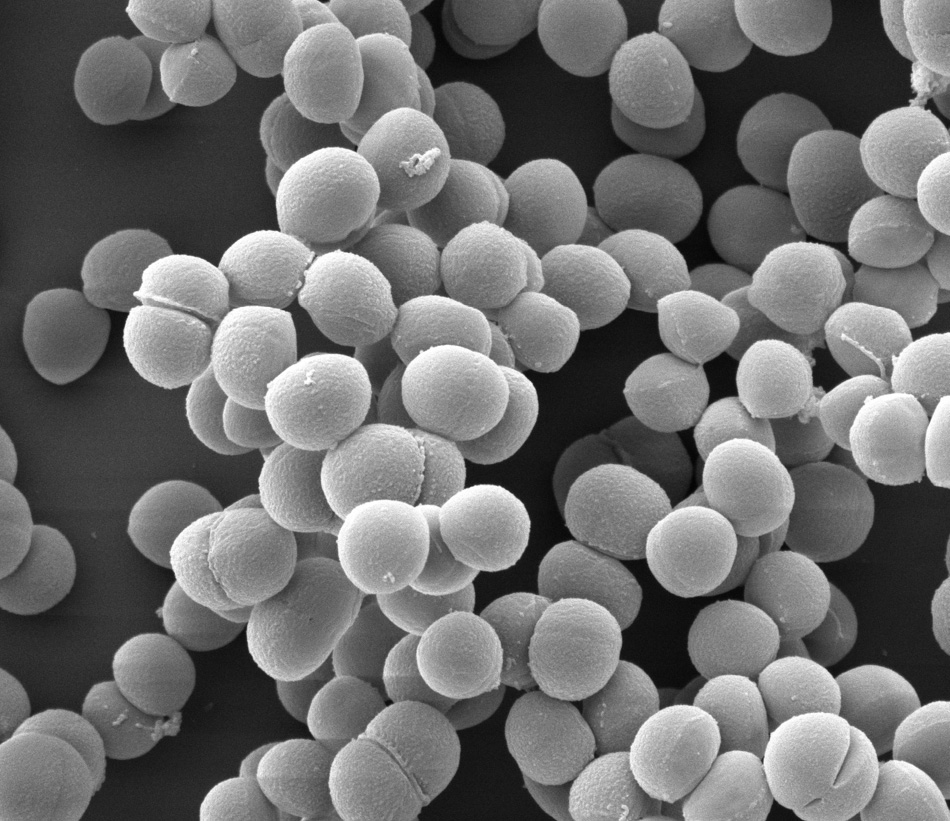
Horizontal gene transfer
in Staphylococcus aureus
A major consequence of horizontal gene transfer in S. aureus has been the emergence of resistance to antibiotics, characterized by difficult to treat methicillin (MRSA) and vancomycin (VISA) infections. S. aureus encodes many mobile genetic elements, such as transposons, prophages, and plasmids that can mediate horizontal gene transfer. However, the low frequency of these events can make them difficult to capture in a laboratory setting. Characterizing how DNA moves in S. aureus will enhance our understanding of how antibiotic resistance and virulence genes disseminate: What molecular mechanisms initiate a gene transfer event? What route does that DNA take to a recipient? How do these recipient cells combat the uptake of foreign DNA?

Adapting phage machinery
to manipulate DNA
Viruses that infect bacteria, phages, and the bacteria-encoded immune systems that defend against them have proven to be a particularly rich resource for molecular biology tools. Phages often encode extremely efficient recombination systems and a small portion of these have been exploited to engineer bacterial genomes. Due to the sheer number of phages, the vast majority of these recombination systems remain unstudied. Developing these recombinases as genome engineering tools will unlock new avenues to efficiently edit prokaryotic and eukaryotic genomes: What systems can be developed with current phage to mediate genome engineering? Can unidentified phage recombinases be harnessed for genome editing?
Images courtesy of Hilda Amalia Pasolli and Christian Baca